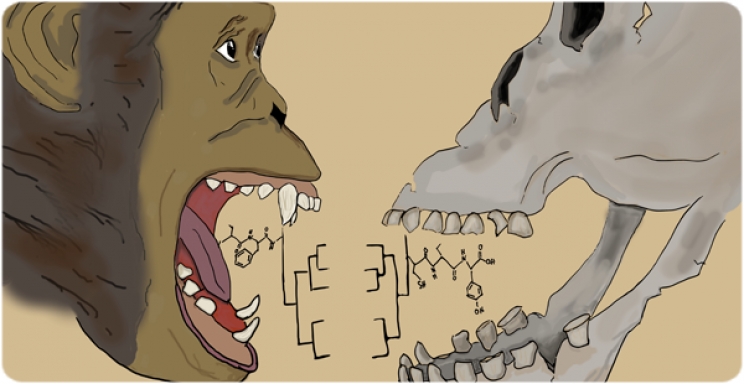
Tooth enamel is remarkably durable, and even after thousands or even millions of years, it retains short protein fragments that can offer clues about ancient species. These proteins, collectively known as the enamelome, have been instrumental in reconstructing evolutionary relationships.
However, since the enamelome is derived from only about 10 genes, and its proteins degrade over time, scientists have debated whether it provides enough information for accurate evolutionary analysis. This study takes a closer look at this question, assessing the reliability of enamel proteins in determining the placement of fossil species within the primate family tree.
The research team, led by Esther Lizano, Junior Distinguished Researcher at the ICP, and Tomàs Marquès-Bonet, Principal Investigator at the Institut de Biologia Evolutiva (IBE) and ICREA Research Professor at Universitat Pompeu Fabra (UPF) and head of the ICP Paleogenomics and Paleoproteomics research group, simulated ancient protein degradation patterns and compared them with known phylogenetic trees based on genomic data.
By analyzing 14 key enamel proteins across 232 primate species, they assessed the capacity of these proteins to reconstruct evolutionary relationships. The results indicate that while the enamelome can place fossil species at least at the family level, its resolution within closely related species remains limited due to the small number of informative loci.
One of the key findings is that the inclusion of certain collagen proteins can introduce phylogenetic errors, as observed in the incorrect placement of tarsiers in relation to other primates. The study highlights the importance of selecting appropriate protein datasets and considering the effects of protein degradation when interpreting ancient proteomic data.
This research has significant implications for paleoproteomics, an emerging field that has enabled scientists to study evolutionary relationships in deep time where ancient DNA is no longer available. The study provides a framework for evaluating the feasibility of future proteomic-based phylogenetic reconstructions and underscores the need for methodological refinements in this area.
This publication represents the first study developed within the collaborative framework of the Paleobiology Unit, ICP-CERCA, following the recent incorporation of the ICP as an Associated Unit of the Consejo Superior de Investigaciones Científicas (CSIC) through IBE.
Main image: Artistic representation of phylogenies derived from fossil tooth enamel proteins, showcasing the potential of paleoproteomics in the study of extinct species. Illustration by Johanna Krueger. (Extracted from the cover image of Genome Biology and Evolution, Vol. 17, No. 2, 2025)
Original article:
Fong-Zazueta, R., Krueger, J., Alba, D. M., Aymerich, X., Beck, R. M. D., Cappellini, E., Carrillo-Martín, G., Cirilli, O., Clark, N., Cornejo, O. E., Farh, K. K.-H., Ferrández-Peral, L., Juan, D., Kelley, J. L., Kuderna, L. F. K., Little, J., Orkin, J. D., Paterson, R. S., Pawar, H., Marques-Bonet, T., & Lizano, E. (2025). Phylogenetic signal in primate tooth enamel proteins and its relevance for paleoproteomics. Genome Biology and Evolution, 17, evaf007. https://doi.org/10.1093/gbe/evaf007